Abstract
Neuroinvasive human herpesvirus 1 (HSV-1) recombinants use intergenomic recombination to diversify viral populations. Research efforts to assess HSV-1 recombination are often complicated by the use of attenuating mutations, which differentiate viral progeny but unduly influence replication and spread. In this work, we generated viruses with markers that allowed sorting of viral progeny with limited attenuation of viral replication.
We isolated viruses, harbouring a cyan (C) or yellow (Y) fluorescent protein (FP) expression cassette inserted at two different locations within the viral genome, to visually quantify recombinant progeny based on plaque fluorescence. We found that FP-tagged genomes had a limited negative effect on viral replication and the production of progeny virions. A co-infection of the two viruses resulted in recombinant progeny dependent on a multiplicity of infection and independent of time post-infection, at a rate similar to previous reports.
Sequential passage of mixed viral populations revealed a limited change in the distribution of parental and recombinant progeny. Interestingly, neuroinvasive propagation within neuronal cultures and an in vivo mouse model revealed large random changes in parental and recombinant distributions in viral populations. In conclusion, our approach highlights the utility of FP-expressing viruses in providing new insights into HSV-1 recombination mechanisms.
Keywords:
alphaherpesvirus, HSV-1, recombination, fluorescent protein, cell-to-cell spread, neuroinvasion, neuron culture, intravitreal injection
Cells and Viruses
Vero cells, a kind gift from the Enquist laboratory, were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM), containing 10% (v/v) fetal bovine serum and 1% penicillin/streptomycin. All steps of virus isolation, analysis and production were completed on Vero cells. All infections were performed with HSV-1 strain 17, which was derived from a sequence-verified isolate provided by the Enquist laboratory.
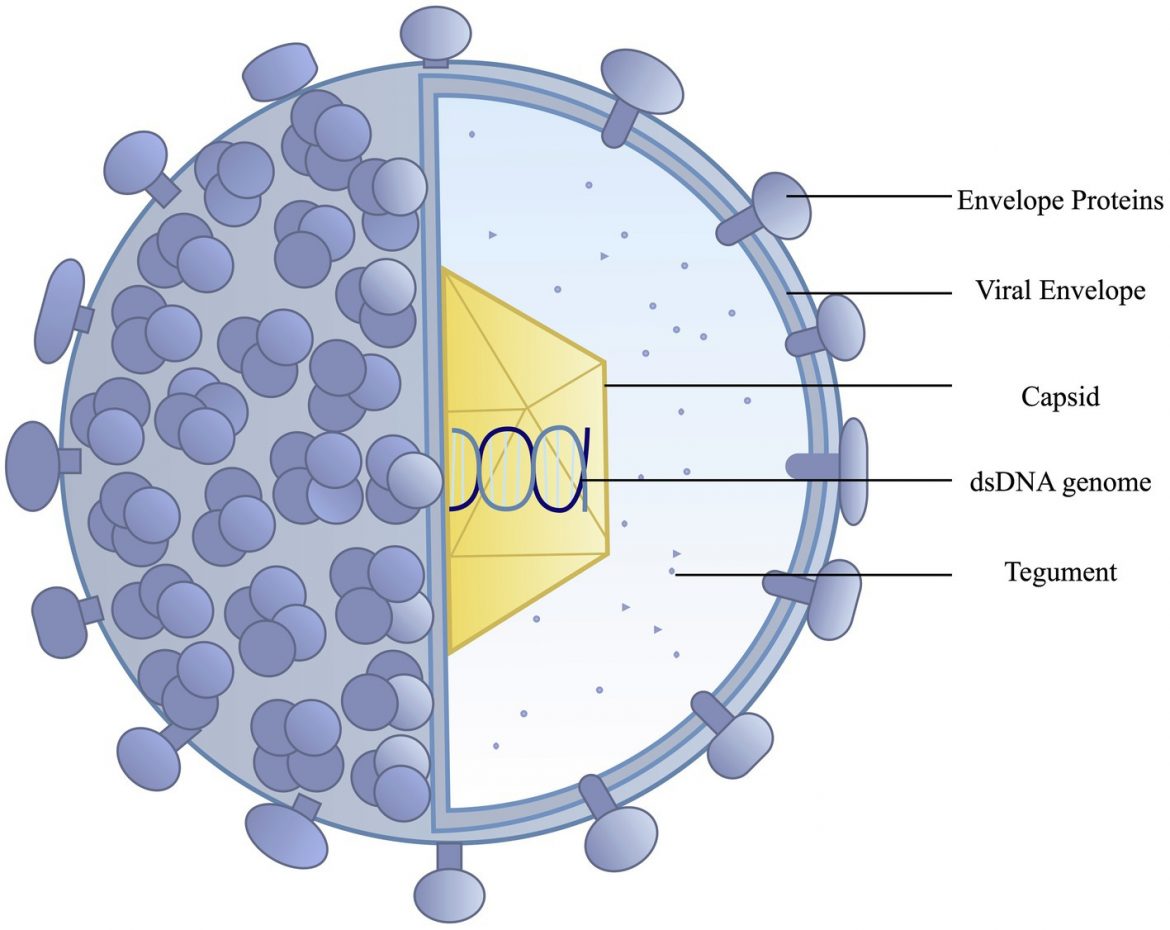
Viruses that were developed for recombination expressing yellow fluorescent protein (YFP) with a nuclear localization sequence (NLS) (HSV-1 OK12 and referred to here in the manuscript as HSV-1 YFPUL ) and cyan fluorescent protein (CFP)-NLS (HSV-1 MT002 referred to as HSV-1 CFPUS in the manuscript), both derived from HSV-1 strain 17.
Construction of viruses expressing FP
HSV-1 YFPUL was a previously described virus that contained a genetic cassette that inserted between the viral UL37/UL38 genes, allowing cytomegalovirus (CMV)-driven expression of a nuclear localization sequence that was tagged as eYFP. HSV-1 CFPUS was a new recombinant strain, which was isolated for this work with a similar genetic cassette inserted between the US1/US2 viral genes, however, this cassette encoded a nuclear localization sequence, marking Turquoise2. Both sites in the viral genome that were targeted for the insertion of this gene cassette had previously been shown to tolerate gene insertions without a reduction in viral titer.
Insertion of the Turquoise2 expression cassette required the construction of a plasmid containing an HSV-1 OK12-like fluorophore expression cassette, which was flanked by homology arms that would promote recombination in the US1/US2 region of the HSV-1 genome. one. Approximately 500 bp of homologous sequence flanking the 5′ and 3′ of the Us 1/2 insertion site was amplified by PCR from purified HSV-1 strain 17 DNA. The Turquoise2 CMV-driven expression cassette was amplified by PCR from the plasmid vector, pMT06. All PCR amplification was performed with a Q5 DNA polymerase mix (Qiagen, Hilden, Germany).
The PCR products were visualized and isolated from a 1% Tris-Acetate EDTA (TAE) agarose gel, followed by purification by gel extraction (Qiagen). All PCR products contained 15 bp of the homologous overlapping sequence to facilitate cloning In-Fusion (ClonTech, Mountain View, CA, USA) into a plasmid backbone containing ampicillin resistance. Generation of the final construct was achieved with a 3-way ligation reaction, followed by transformation and isolation of resistant bacterial colonies. Positive recombinant clones were identified by colony PCR. The resulting isolated and purified plasmid was verified by Sanger sequencing.
To isolate HSV-1 viruses with inserts at the US1/US2 locus, purified plasmids were linearized with a double XmnI/XbaI digest. The linearized plasmid was co-transfected with purified HSV-1 strain 17 DNA into Vero cells. The resulting progeny plaques were screened for Turquoise2 expression, followed by three subsequent rounds of plaque isolation to obtain high-quality, stable, pure isolates. All viral stocks were verified by fluorescence microscopy for maintenance of FP expression. A minimum of 100 plaques were imaged to ensure greater than 99% homogeneity of expression across the population of viruses used for infections.
Microscopy
All fluorescent images were performed on a TiEclipse inverted fluorescence microscope (Nikon Instruments, Melville, NY, USA), which employed a motorized objective turret, motorized X-Y stage, and mechanical shutters for transmitted and fluorescent illumination. Fluorescence illumination was supplied by a SpectraX illuminator (Lumencor, Beaverton, OR, USA), which contained six high-intensity bandpass-restricted LED illuminators that were oriented for a single illumination path.
The excitation sources were combined with specific multipass dichroic mirrors and emission filters, including CFP/YFP/RFP filter pairs (Chroma Technology Corp., Bellows Falls, VT, USA). To quantify FP expression and frequency of phenotypes in progeny viral populations, a mosaic of multiple images of viral plaque assays was acquired. For each image area, three channels were captured, which acquired a transmitted light, CFP and YFP.
Recombination Quantification
To assess the frequency of recombination that occurs during HSV-1 replication and propagation, monolayers of Vero cells were inoculated with equal amounts of HSV-1 CFPUS and YFPUL at the indicated multiplicity of infection (MOI). After one hour of inoculation at 37°C, the monolayer was washed with phosphate-buffered saline. Cells and supernatant containing progeny virions were harvested by scraping at the indicated times in each experiment after infection.
Infected samples were then subjected to a cycle of freezing and thawing, sonicated, and titer was assessed by plaque assay. The resulting plaques were imaged for FP expression from individual plaques. Plaques that fluoresced in a single colour were denoted as an original genome and those that fluoresced in two colours, or those with no detectable fluorescence, were counted as recombinant progeny.
The influence of multicycle replication on HSV recombination was determined by the passage of virion progeny from cells co-infected with the two FP-expressing viruses. Progeny from a monolayer of Vero cells co-inoculated at an MOI of 10, of HSV-1 CFPUS and YFPUL, were harvested as described above. Progeny were plated at limiting dilutions on cell monolayers to determine viral titer and quantitate marker expression on plates. This progeny, called the inoculum, was then used to infect Vero cells at an MOI of 10.
The progeny resulting from this first, and subsequently second and third, rounds of passage (a total of three sequential rounds of infection and amplification) were analyzed. . for viral titer and fluorescent examination to quantify the distribution of plaque phenotypes. Statistical analysis of the resulting plaque phenotype distributions was performed in GraphPad Prism 7.0 software (GraphPad Software, La Jolla, CA, USA).
Compartmentalized Neural Cultures
Neuronal cultures were constructed in a three-chamber design, as previously described [23, 24]. Briefly, mouse primary superior cervical ganglia (SCG) were dissected from embryos, which were harvested on days 13.5 to 14.5 from pregnant C57Bl/6 mice. Plastic tissue culture dishes were coated with polyornithine (Sigma-Aldrich, St. Louis, MO, USA), followed by murine laminin (ThermoFisher Scientific, San Francisco, CA, USA), before culture.
Parallel grooves were etched on the surface of culture plates and covered with 1% methylcellulose in 1x DMEM. Autoclaved vacuum grease was applied to one side of CAMP320 Teflon three-compartment rings (Tyler Research, Edmonton, Alberta, CA, USA). The Teflon rings were then placed in the tissue culture dishes so that the parallel grooves were perpendicular and extended along all sides.